We explore the entire tree of life on the research vessel MARIA S. MERIAN: animals, algae, bacteria, and viruses. The cruise focuses on the food web and ocean biodiversity. However, some animals, especially hunters such as squid and whales, are too big, fast or camera-shy for our instruments. To detect these top predators, we search for and collect genetic traces, so to speak, working as zoological forensic scientists. Ocean animals constantly lose cells and DNA when swimming, breathing, eating and defecating, resulting in the release of genetic material. We collect and identify this so-called environmental DNA (eDNA for short) using a method specifically developed to analyse DNA traces collected from environmental samples. Specific DNA sections, such as ribosomal DNA or genes essential for energy production, are analysed and exist in all animals. The same or similar essential genes are used for bacteria or unicellular organisms. These genes are later amplified from the samples via PCR and sequenced via next-generation sequencing technologies. The results are later compared to genetic databases to discover which species occurred in the environmental sample.

Selfie while filtering eDNA water samples. Photo: Babett Günther
So, while our colleagues filter water samples to study phytoplankton and microplankton, we filter water from the CTD in parallel and analyse the DNA later in Kiel in the GEOMAR laboratories. Based on the specific DNA traces, we can determine the presence of certain species, such as the deep-sea cephalopods Histioteuthis and Heteroteuthis. Water sampled every 100 m from the surface down to the seafloor can show us at what depths cephalopods occur and allows us to predict what species of prey predatory whales may encounter during their dives. For this, we work together with cetacean specialists. At the same time, we also analyse the biodiversity of other organisms in the samples, which we could then compare to predict the prey of the cephalopods. Ultimately, we can also compare our genetic results with the distribution and abundance of the diverse species of plankton and animals identified by the MSM126 team members, each of whom has their own taxonomic speciality.
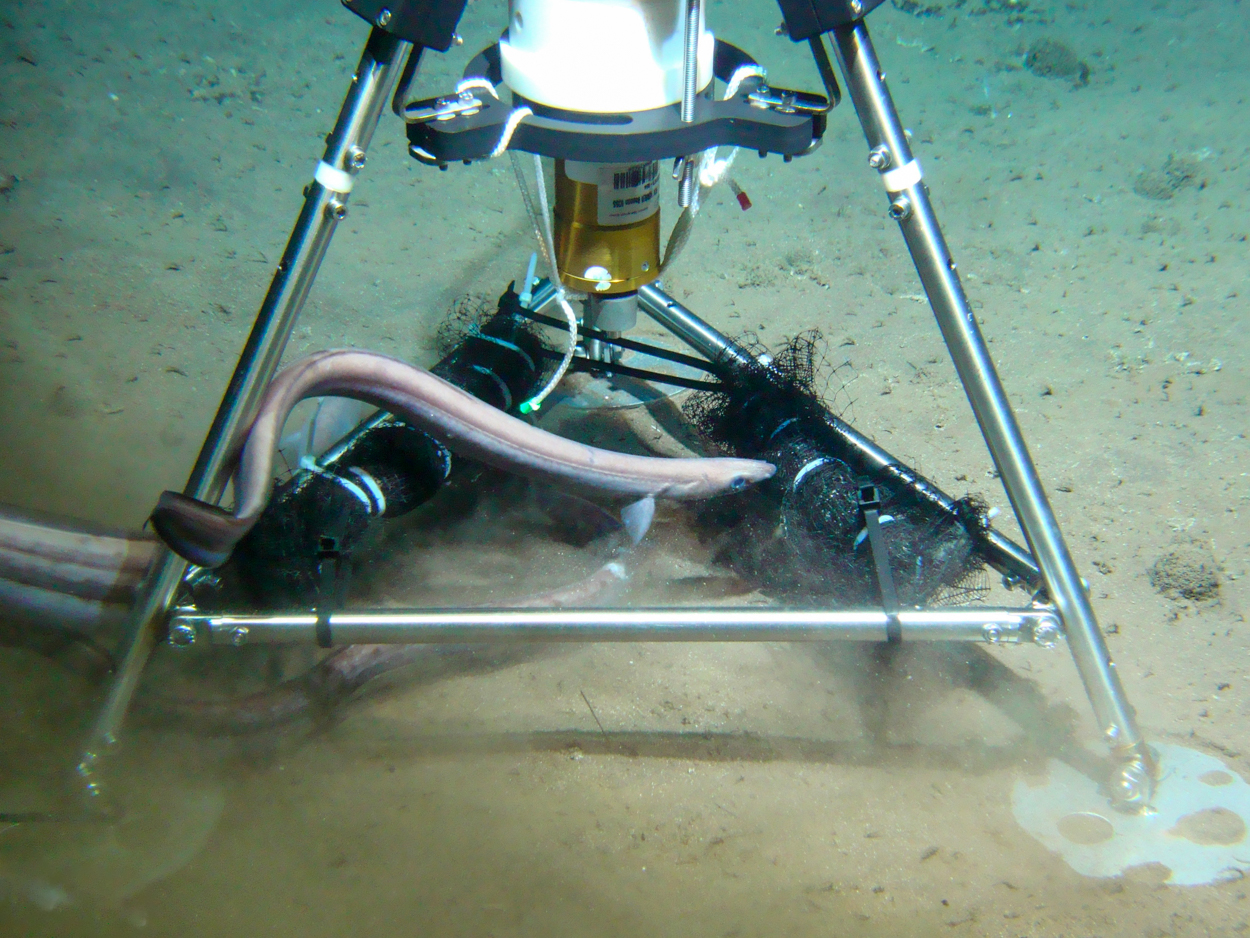
We are also carrying out an innovative eDNA experiment on the deep seafloor during our expedition. For this, the deep-sea robot or ROV has deployed a deep-sea lander with bait on the seafloor to mimic the deposition of a carcass from the overlying water column. This carcass attracts and nourishes many organisms since deep-sea bottom organisms largely depend on food from the overlying water column. We returned to this site several times with the ROV robot to take sediment cores and water samples. These samples are immediately preserved on board for future eDNA analysis. The experiment will investigate 1) which species are attracted to the food deposition event and 2) to what extent the biodiversity and ecosystem of animals and bacteria change over time after the food deposition event.
Environmental genetics, the search for traces in the sea, is a modern addition that supports various classical approaches in marine research and contributes new information and perspectives. It is, therefore, also an essential part of MSM126 to combine research from different disciplines and gain new insights.
Greetings from on board RV MARIA S. MERIAN,
Henk-Jan Hoving and Babett Günther

